Prostate Cancer Detection
A Freeze/Unfreeze Transfer Learning Approach Using Multiple Body Parts
Julien Clément, Johan Jobin, Giuseppe Cuccu, Akansha Bhardwaj and Philipp Cudré-Mauroux
Abstract
Automating the detection of cancer participates to an an early detection and treatment, which increases the chances of recovery. Recent algorithms in artificial intelligence relying on deep learning have shown promising results in this field. Indeed, the usefulness of convolutional neural networks (CNNs) for segmentation or classification tasks is no longer to be proven. However, the performance of these models is often limited by the amount of available data to train the algorithm.
This work first presents a state-of-the-art convolutional neural network for prostate lesion classification. All steps from the data processing to the smallest details regarding the neural network training are explained, ensuring a complete reproducibility of the experiment. This model was evaluated on the official SPIE-AAPM-NCI Prostate MR Classification Challenge dataset and achieved an AUC of 0.76. This result constitutes a solid baseline and confirms the correct functioning of the implementation.
On top of this implementation, a new transfer learning approach using lesions of multiple body parts (brain and lung) was built. This method shows that integrating information from diverse datasets improves automated prostate cancer diagnosis. Indeed, it appears that lesions of these different types of cancer share low-level features that can be used to increase the generalization ability and performance of the prostate lesion classifier. This technique provides a concrete solution to the lack of available data for prostate classification and suggests that many other types of cancers can take advantage of it.
Research paper reproduction
Based on Yang Song et al. “Computer-aided diagnosis of prostate cancer using a deep convolutional neural network from multiparametric MRI: PCa Classification Using CNN From mp-MRI”. en. In: Journal of Magnetic Resonance Imaging 48.6 (Dec. 2018), pp. 1570–1577. ISSN: 10531807.
Process overview
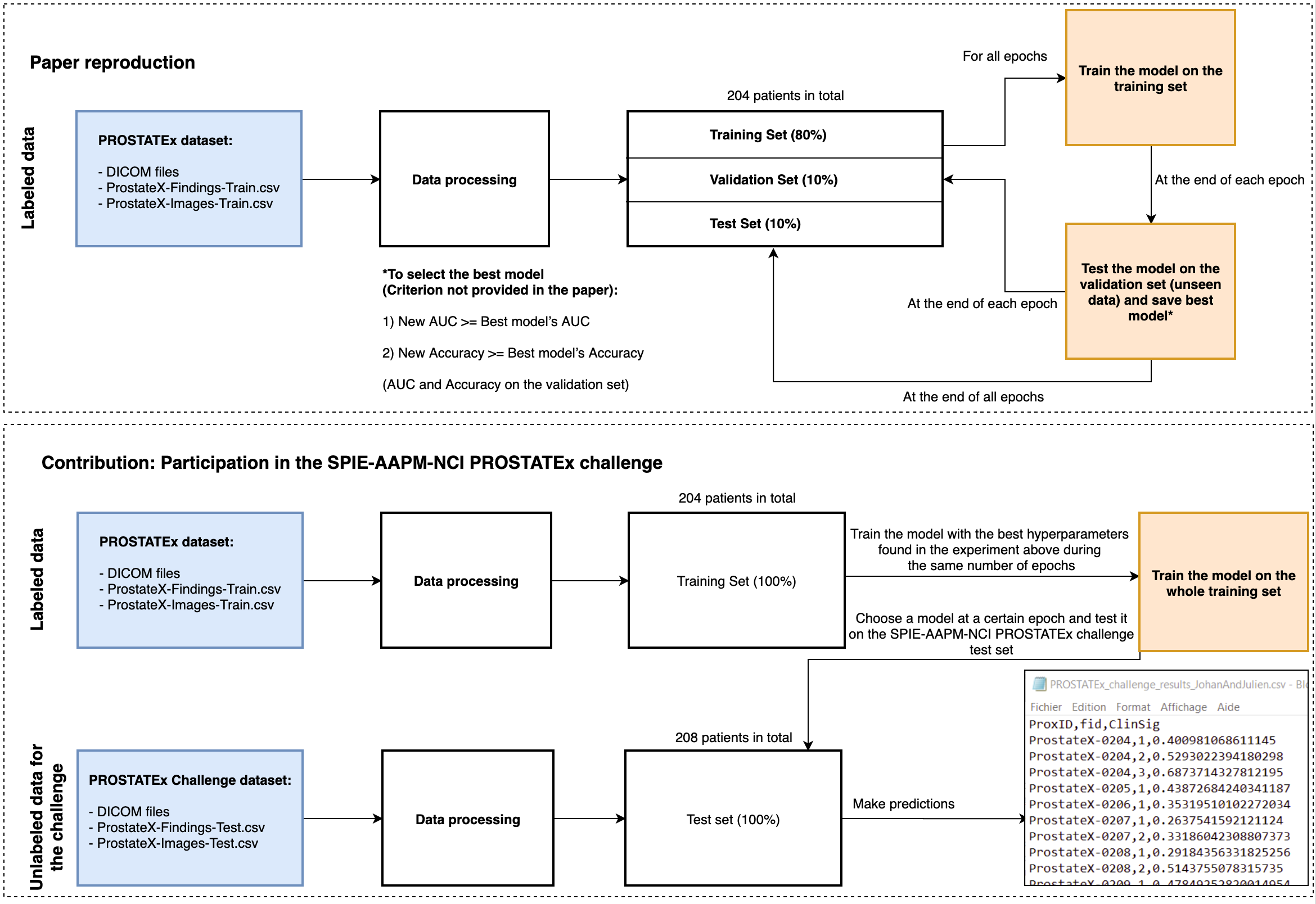
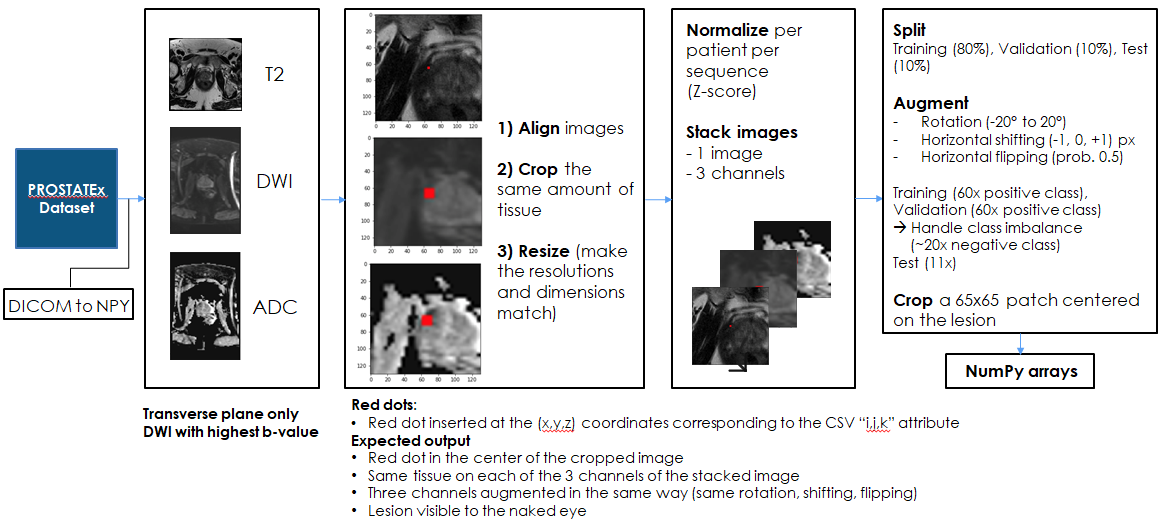
Data processing
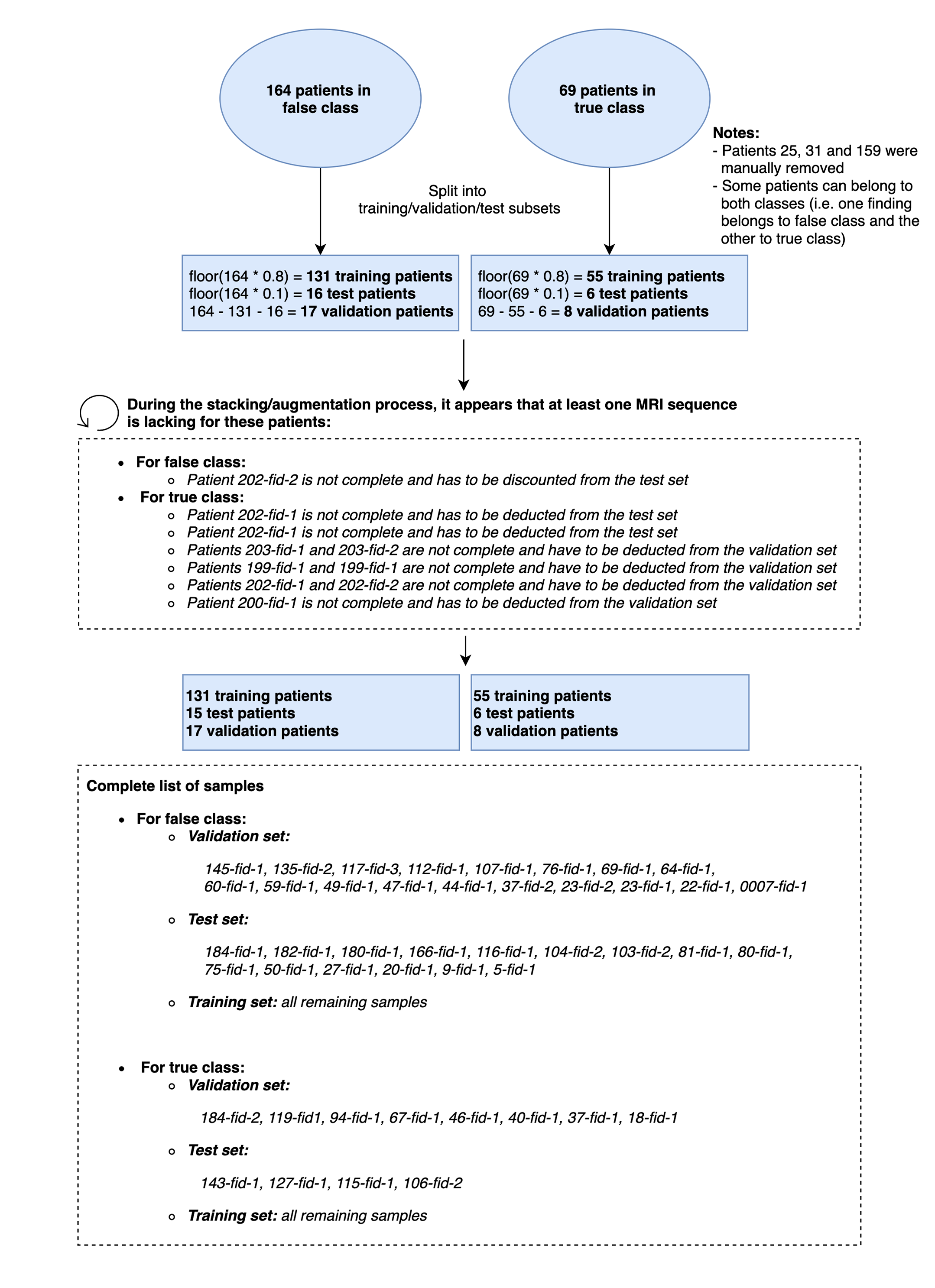
Data splitting
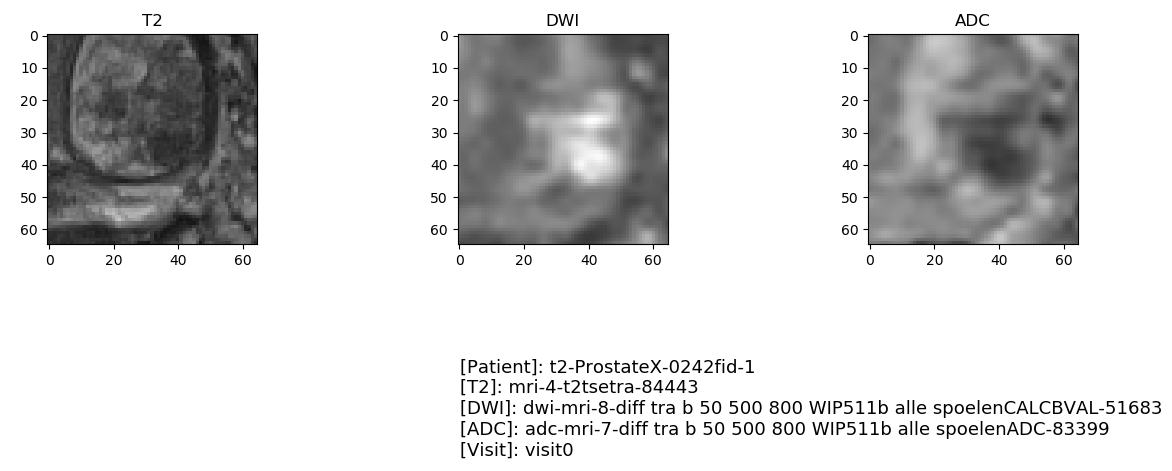
Visual checking
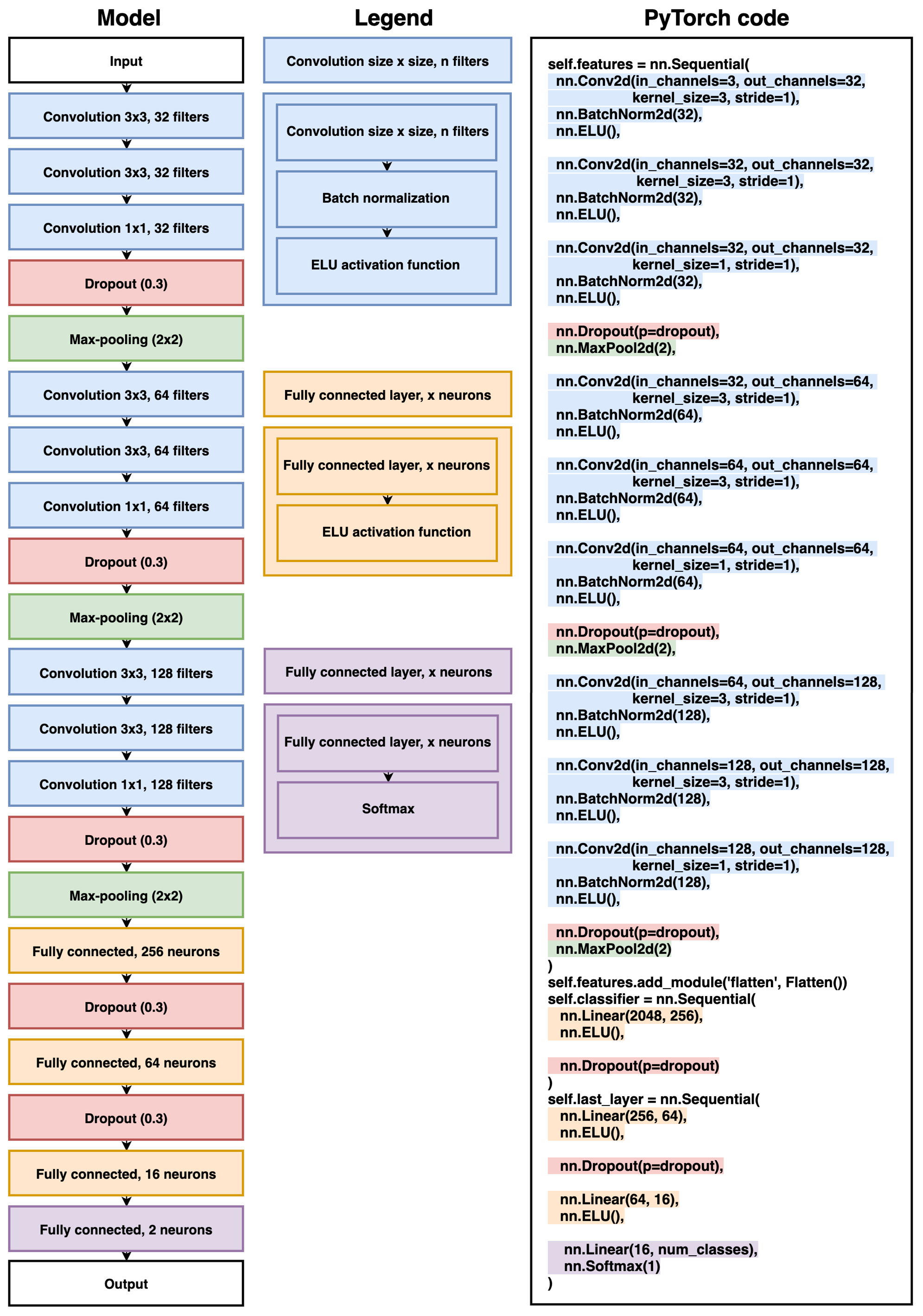
Model architecture
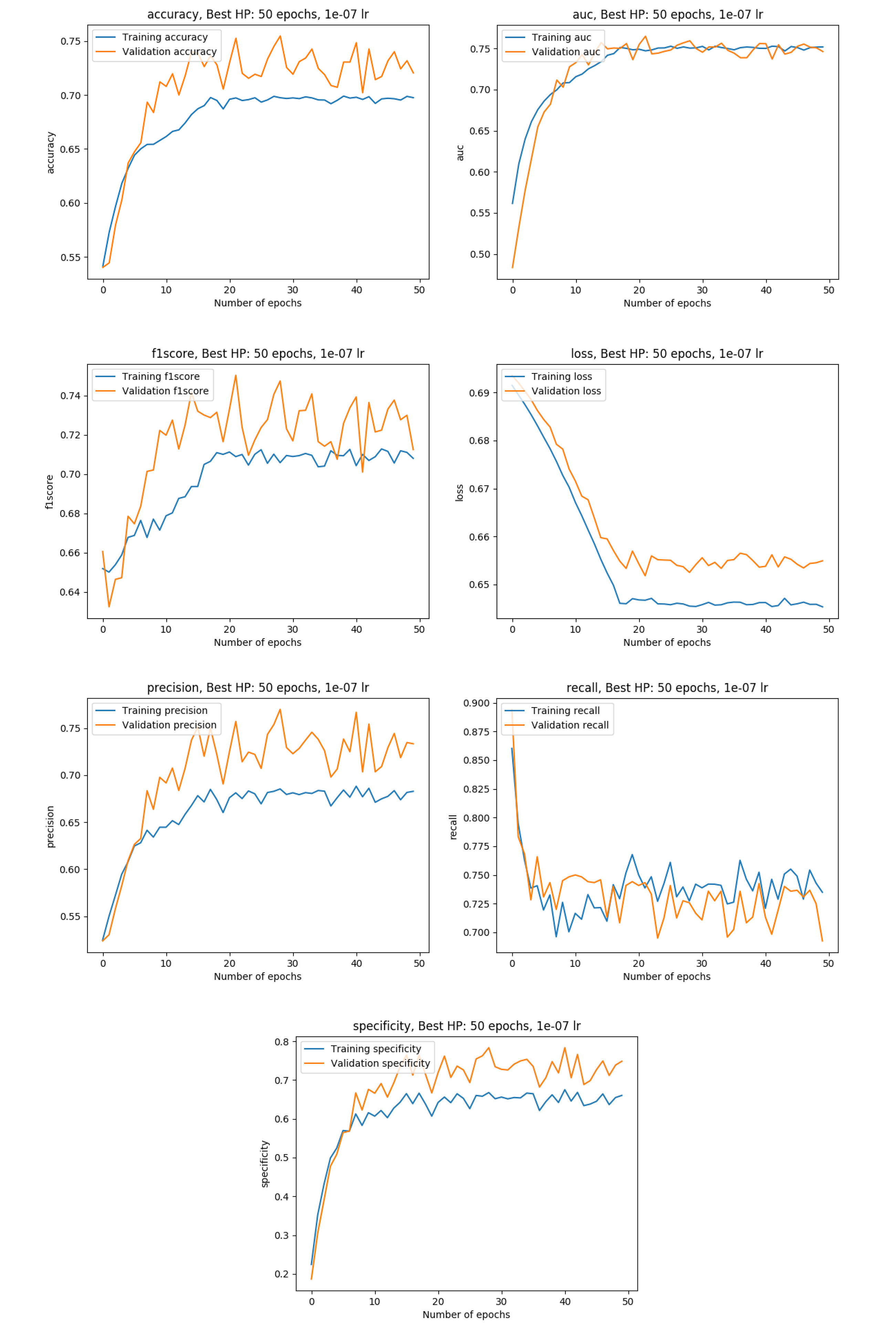
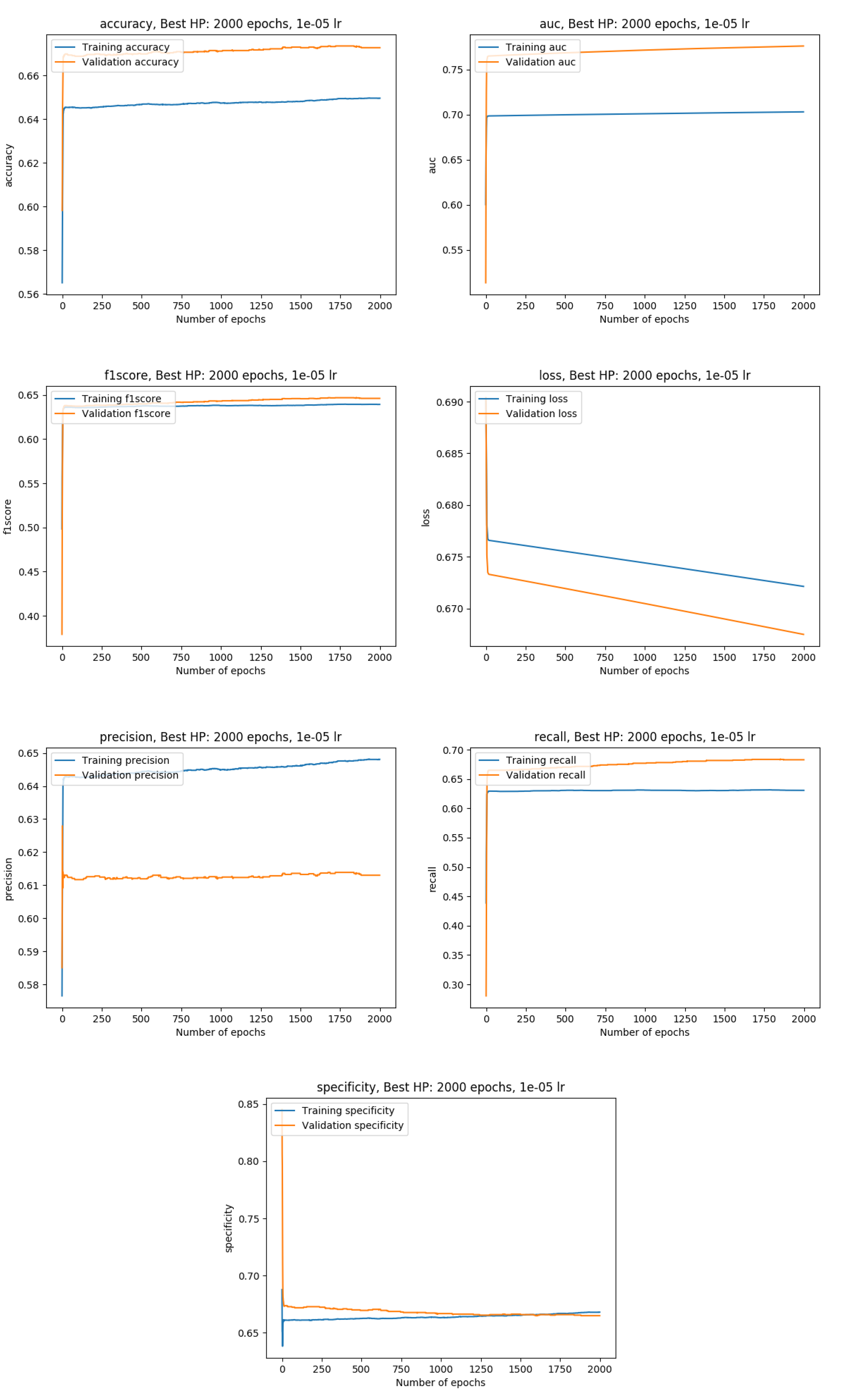
Training metrics with the data split into a training (80%), a validation (10%) and a test set( 10%)
Results of the model on our test set

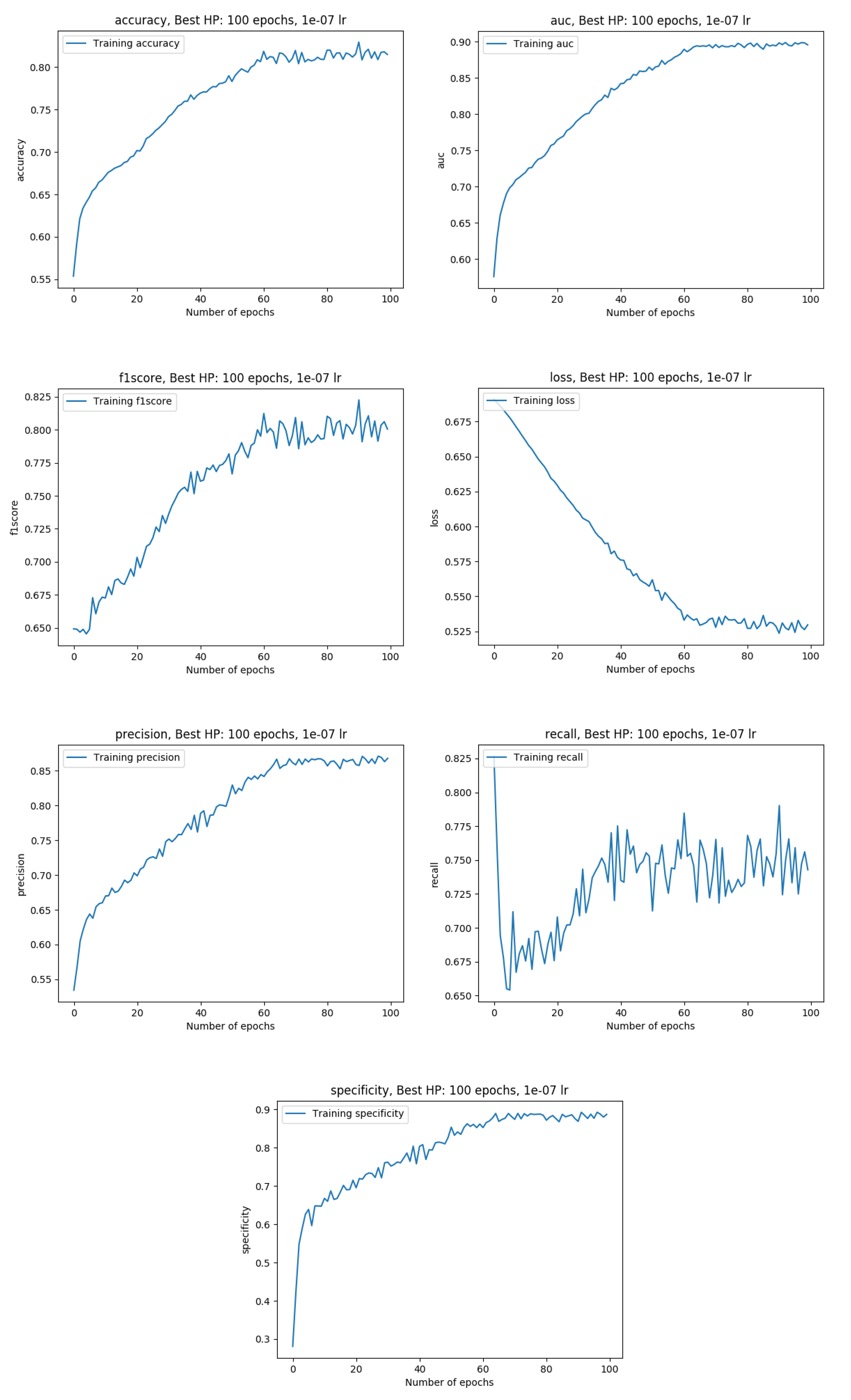
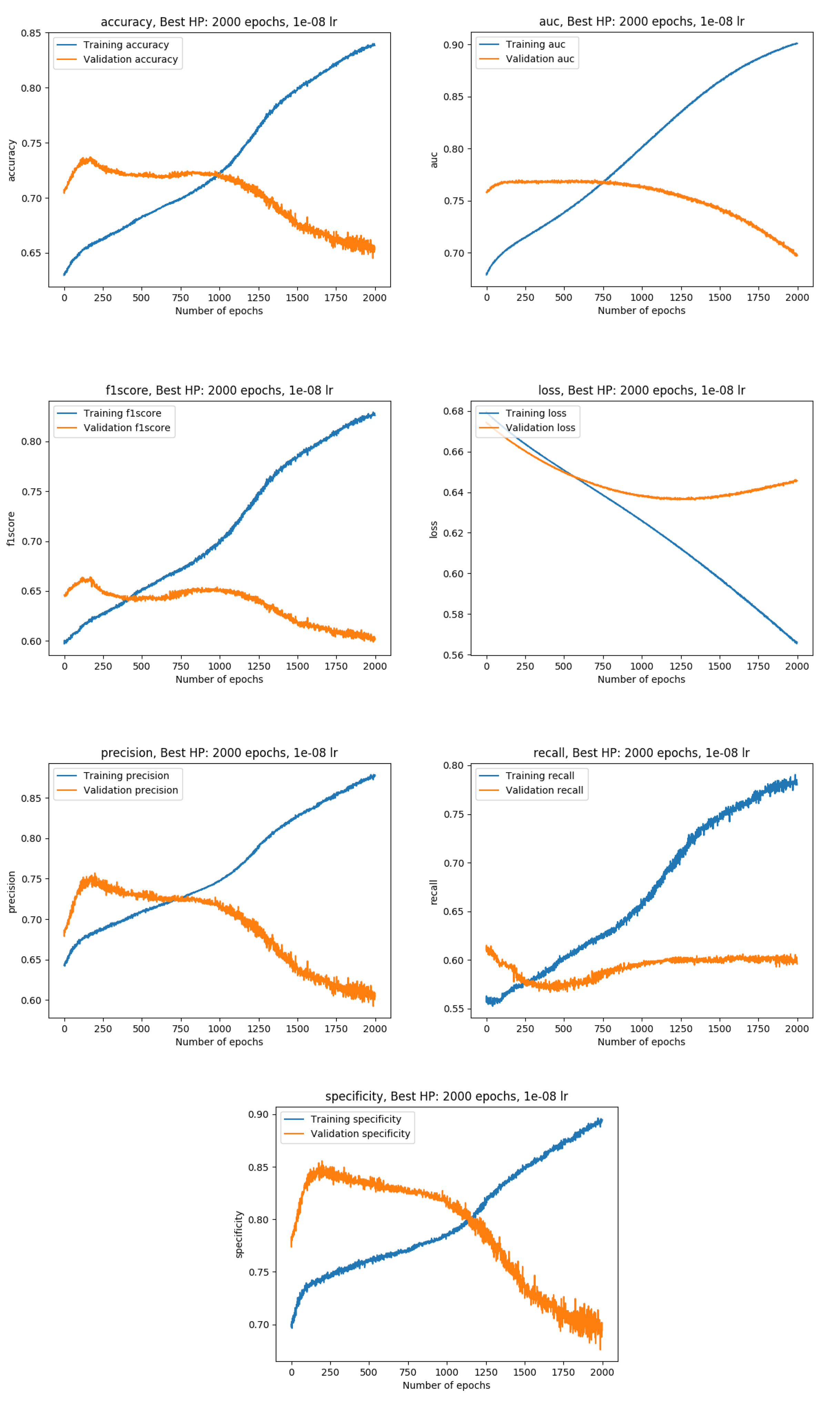
Training metrics with the whole data available (100%)
PROSTATEx challenge result with the model saved at epoch 21:
Freeze/Unfreeze transfer learning using multiple body parts (Prostate, Brain, Lung)
Main idea: split the model into a features extractor and a decision maker
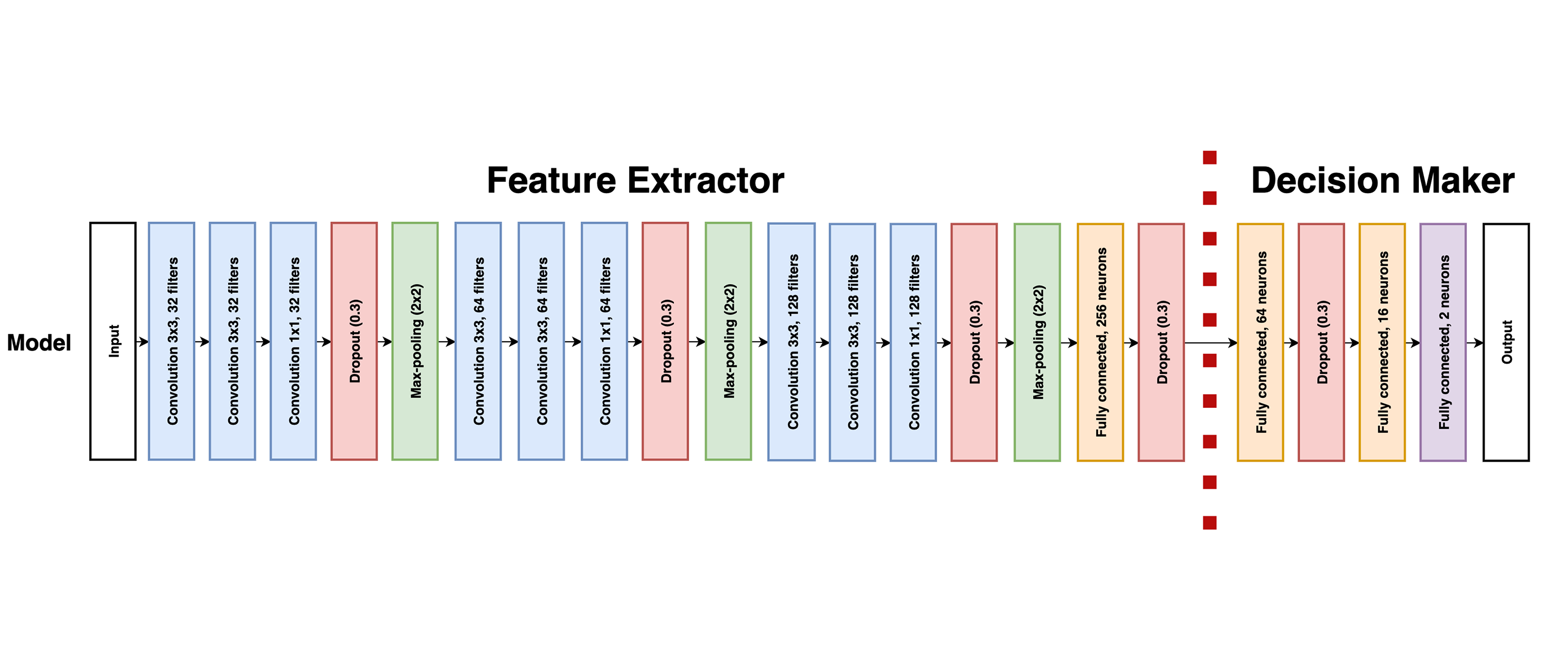
Process overview
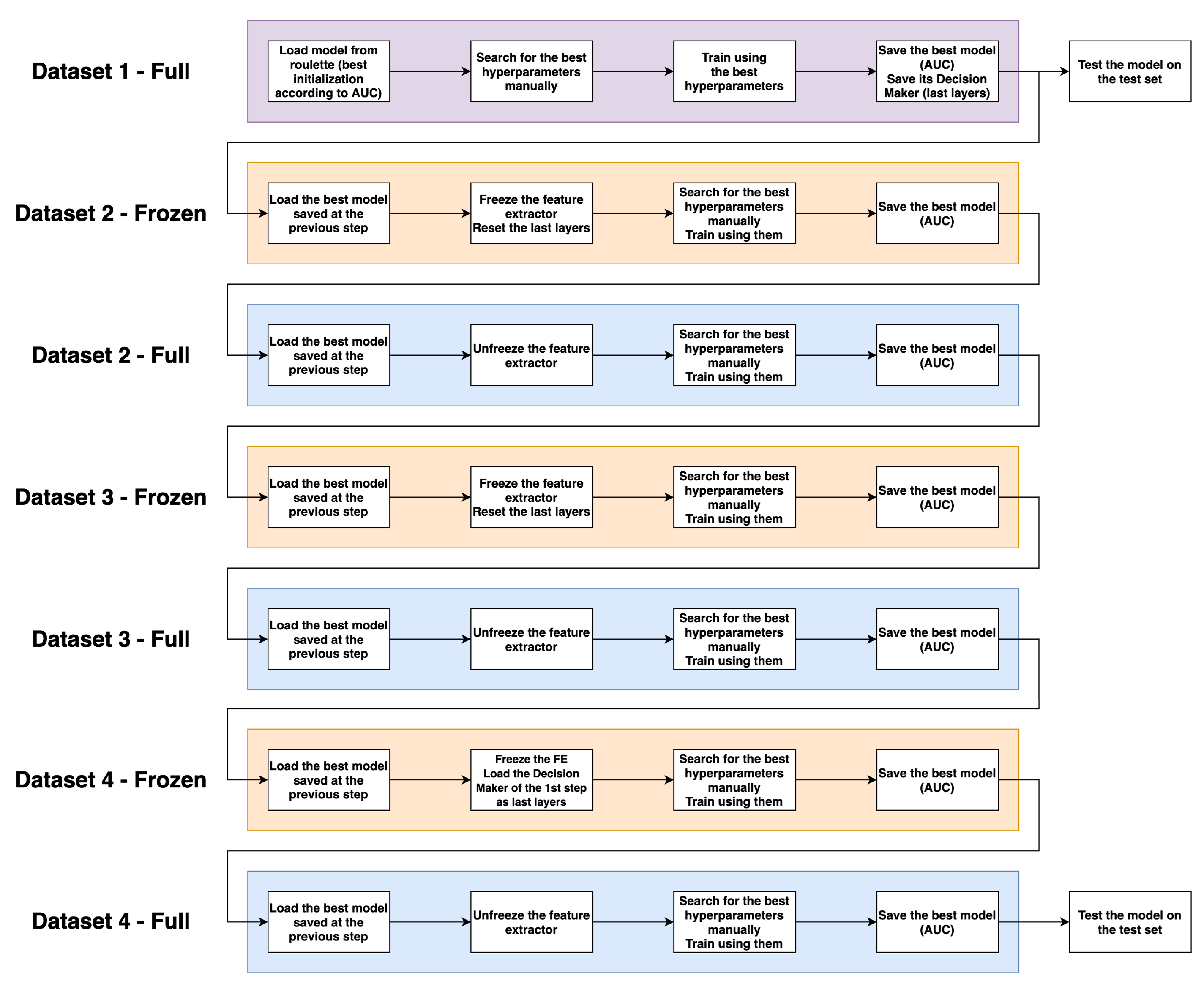
PROSTATEx processing
Lung CT Challenge processing
Kaggle Brain processing
Training results
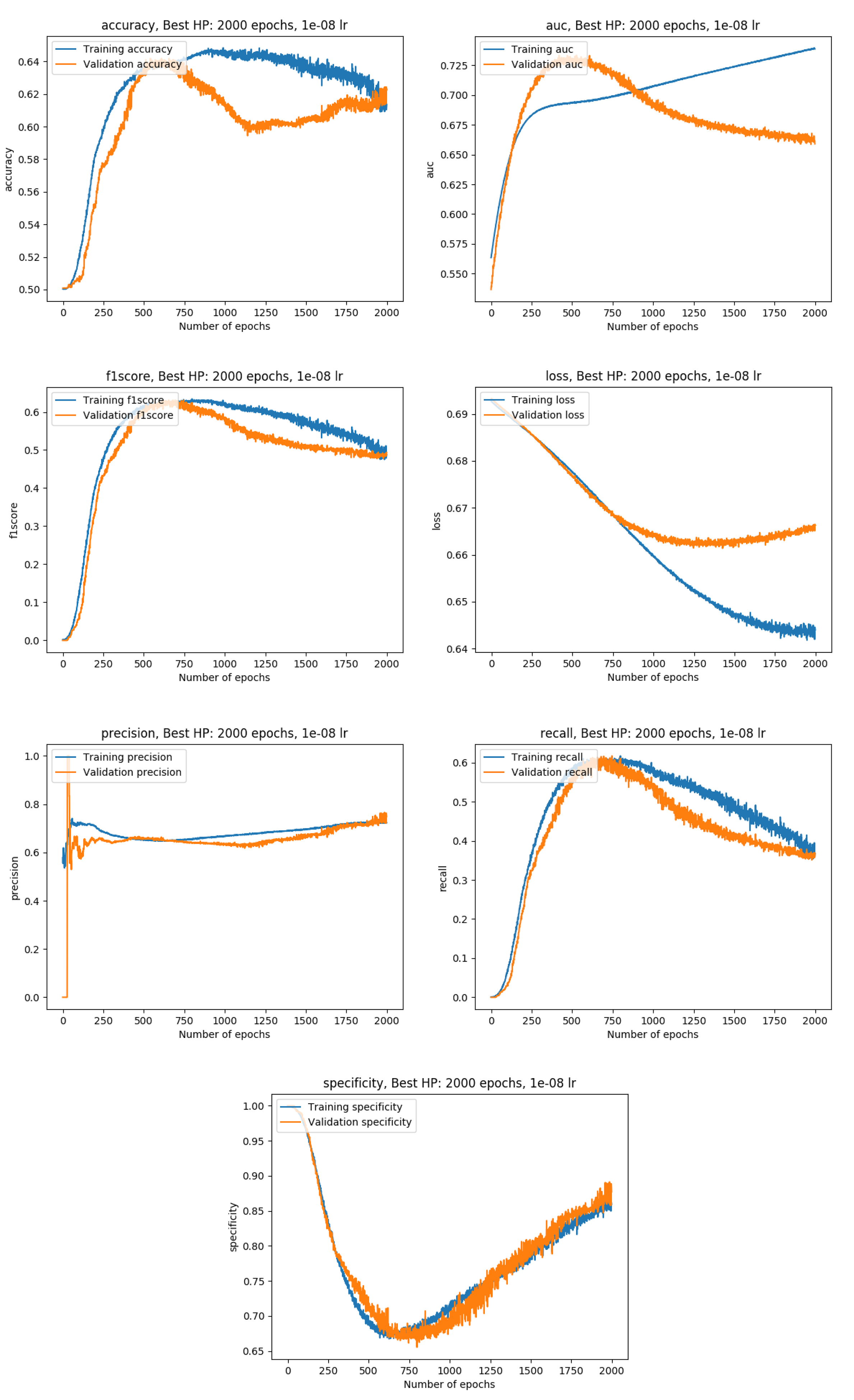
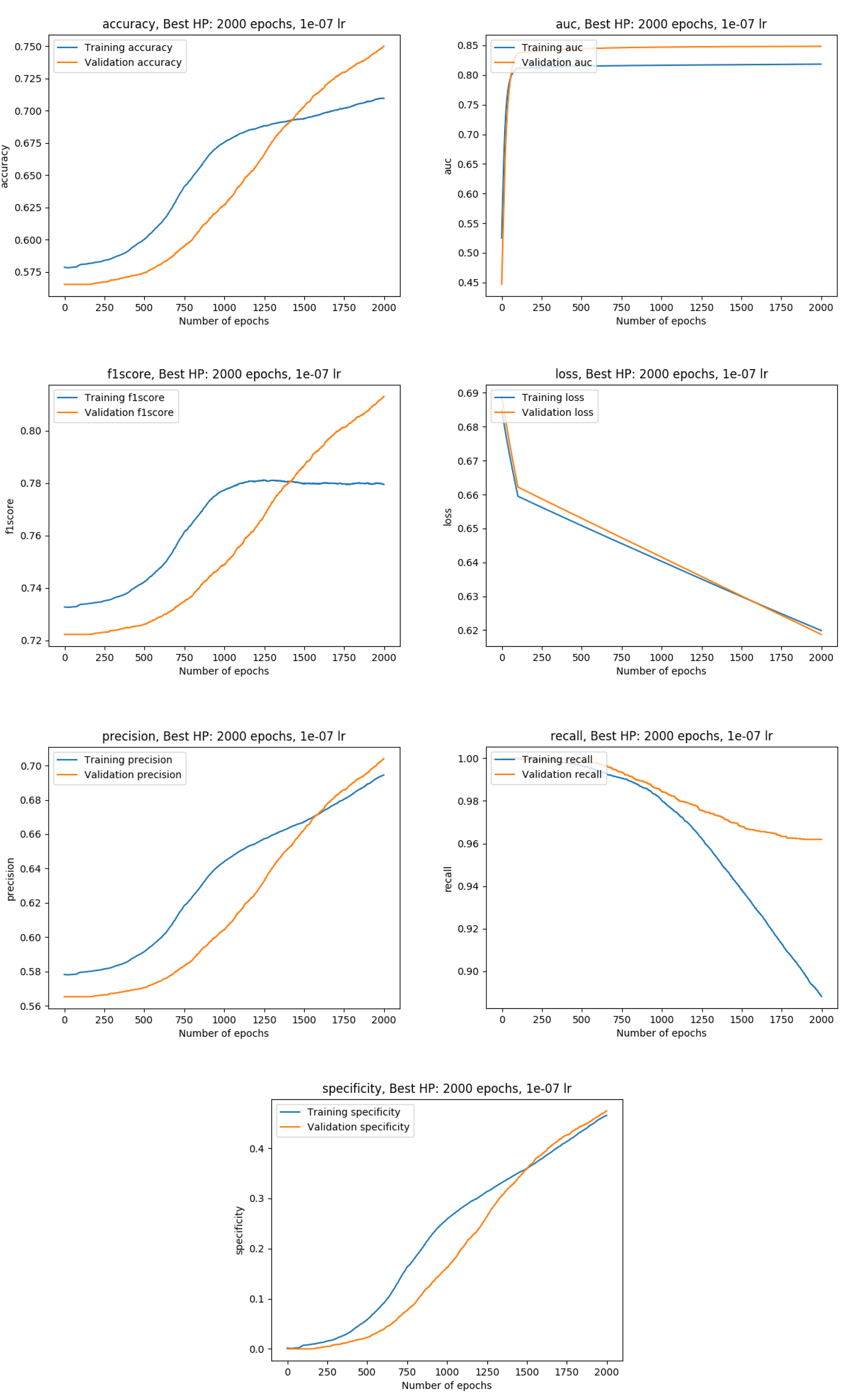
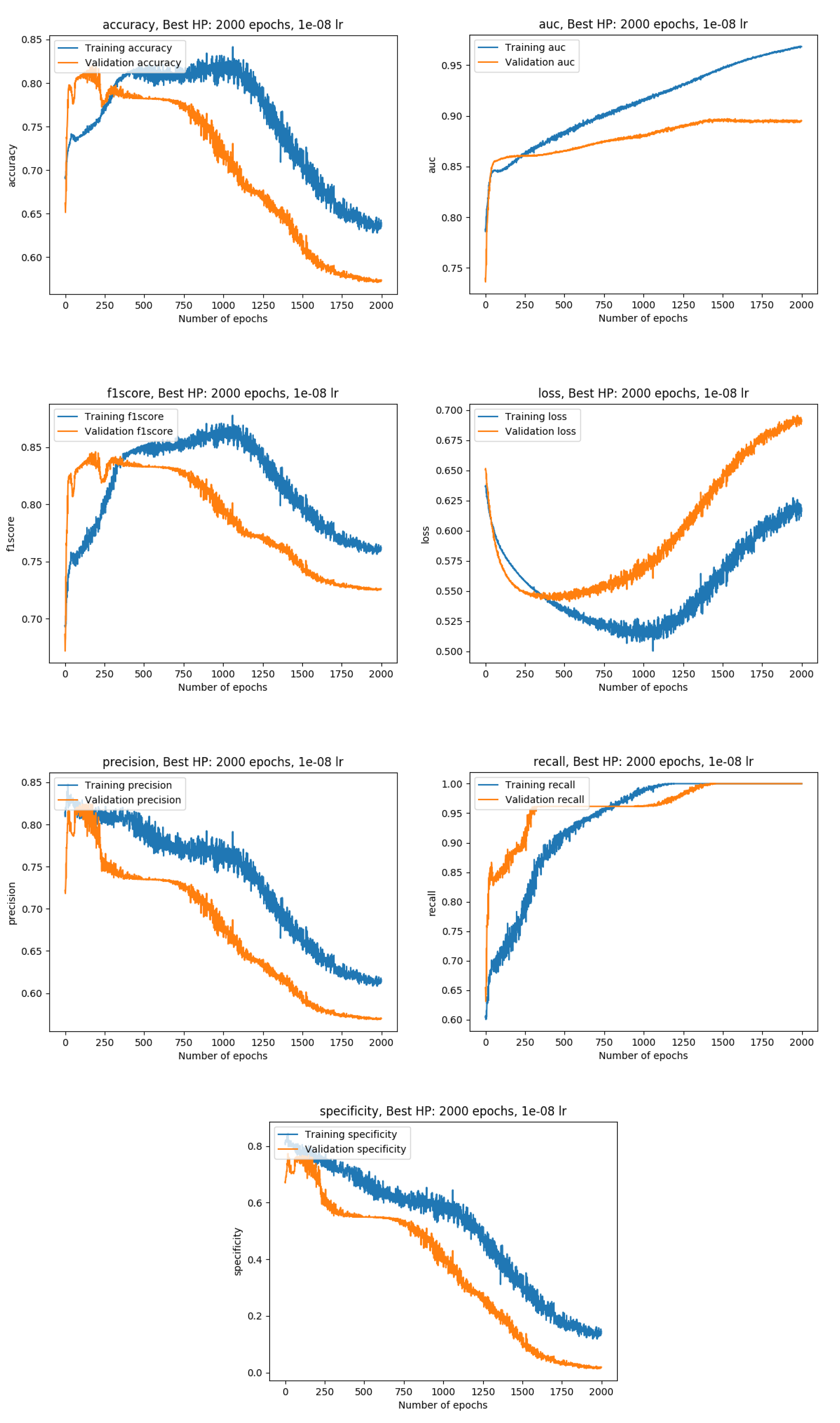
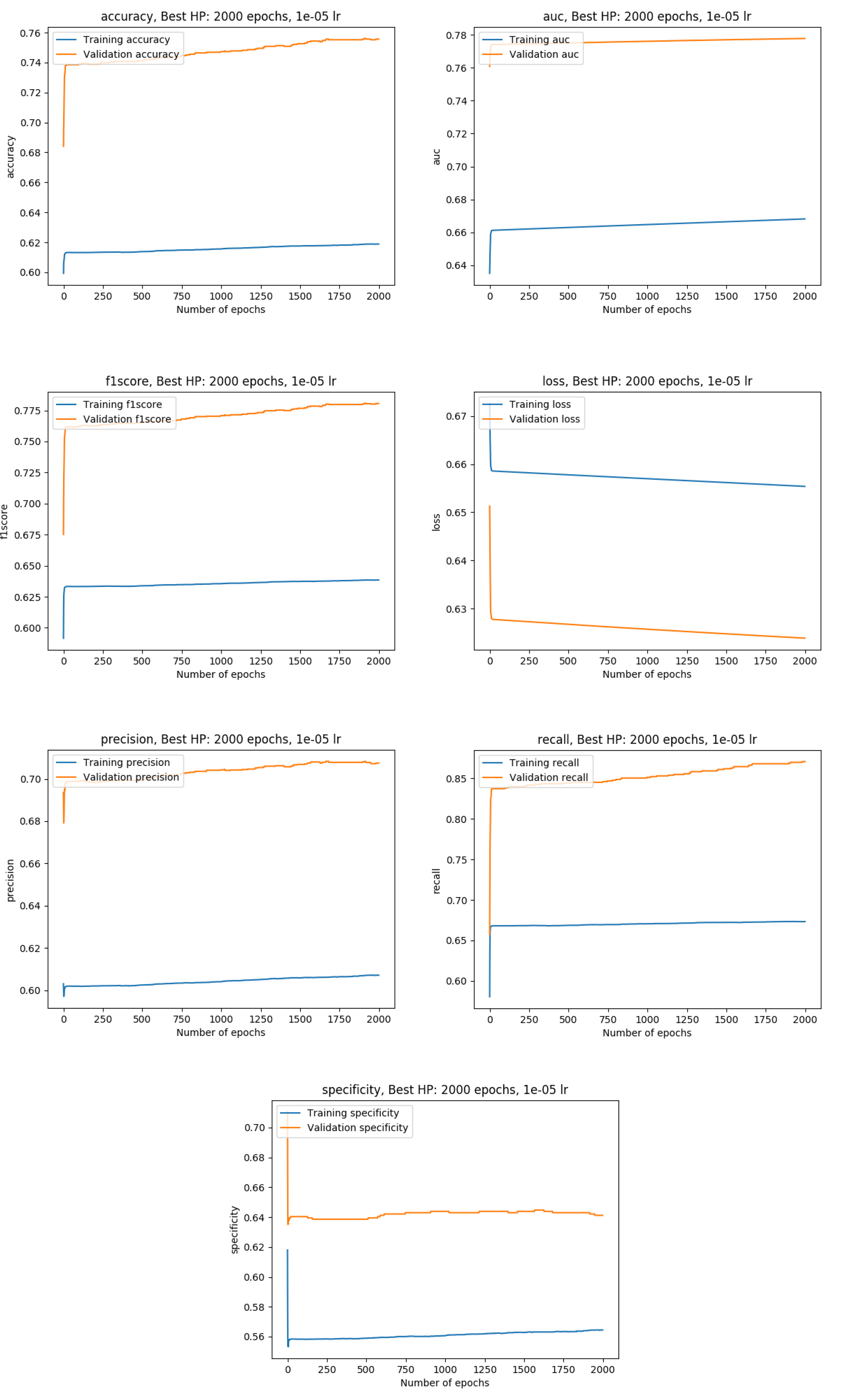
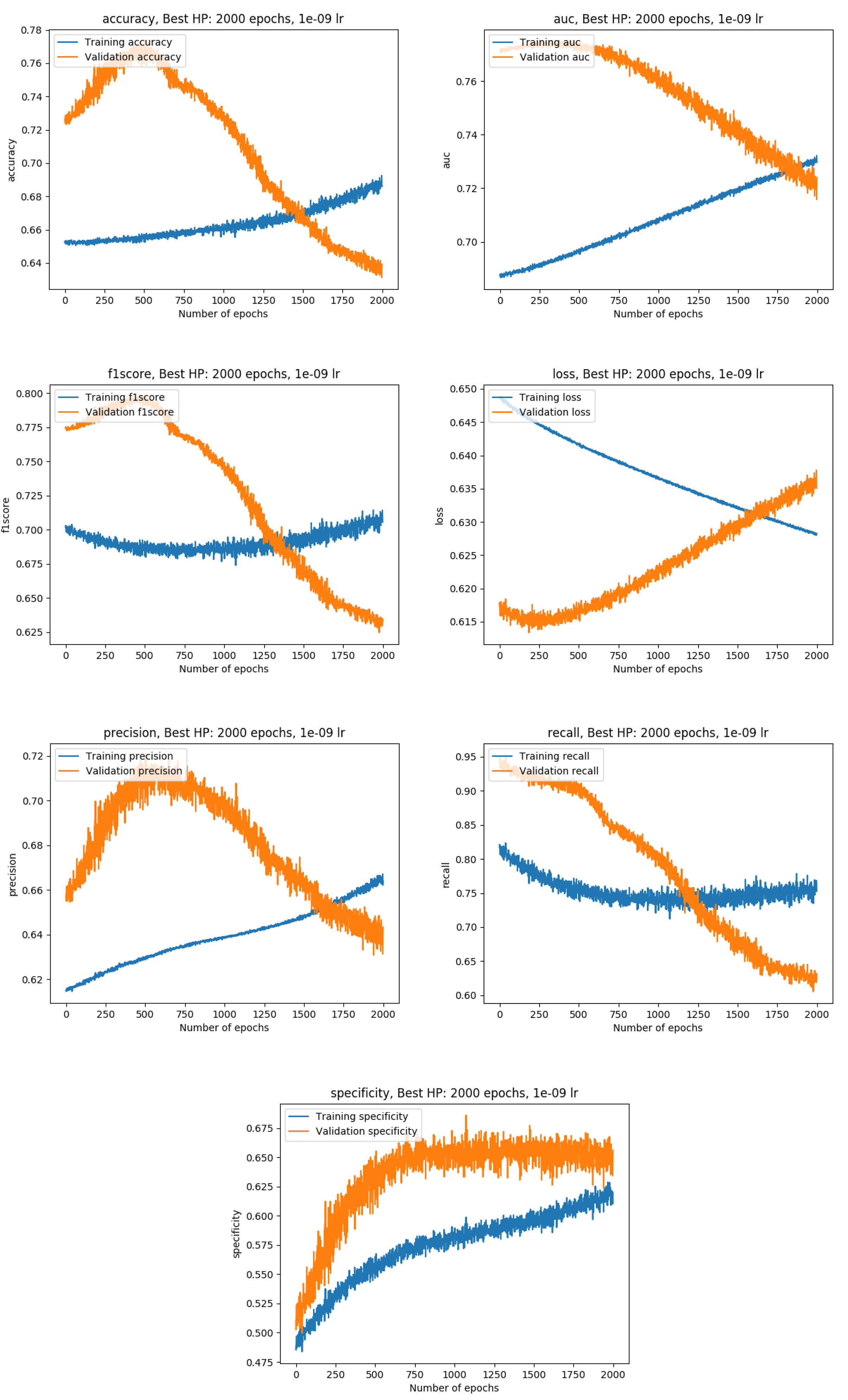
Exact metrics
Use the decision maker trained on the first dataset (PROSTATEx) on all models to quantify the usefulness of brain and lung features for prostate classification
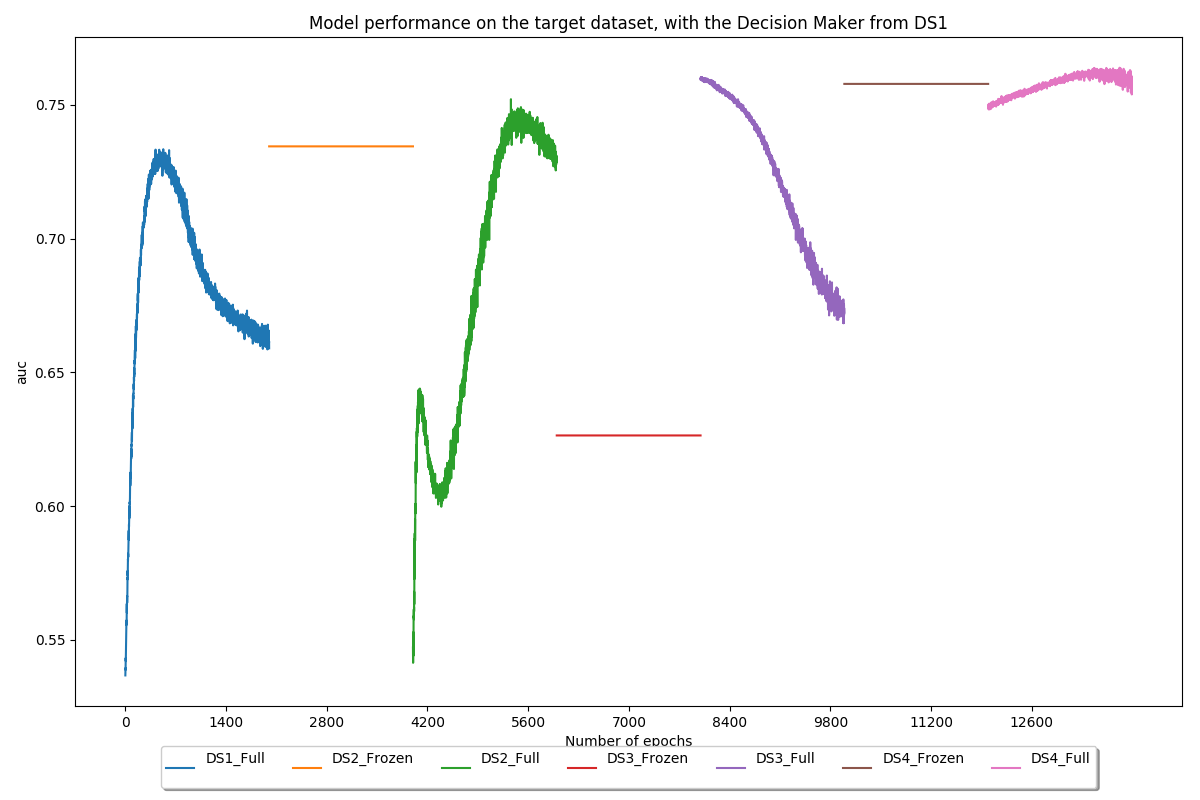
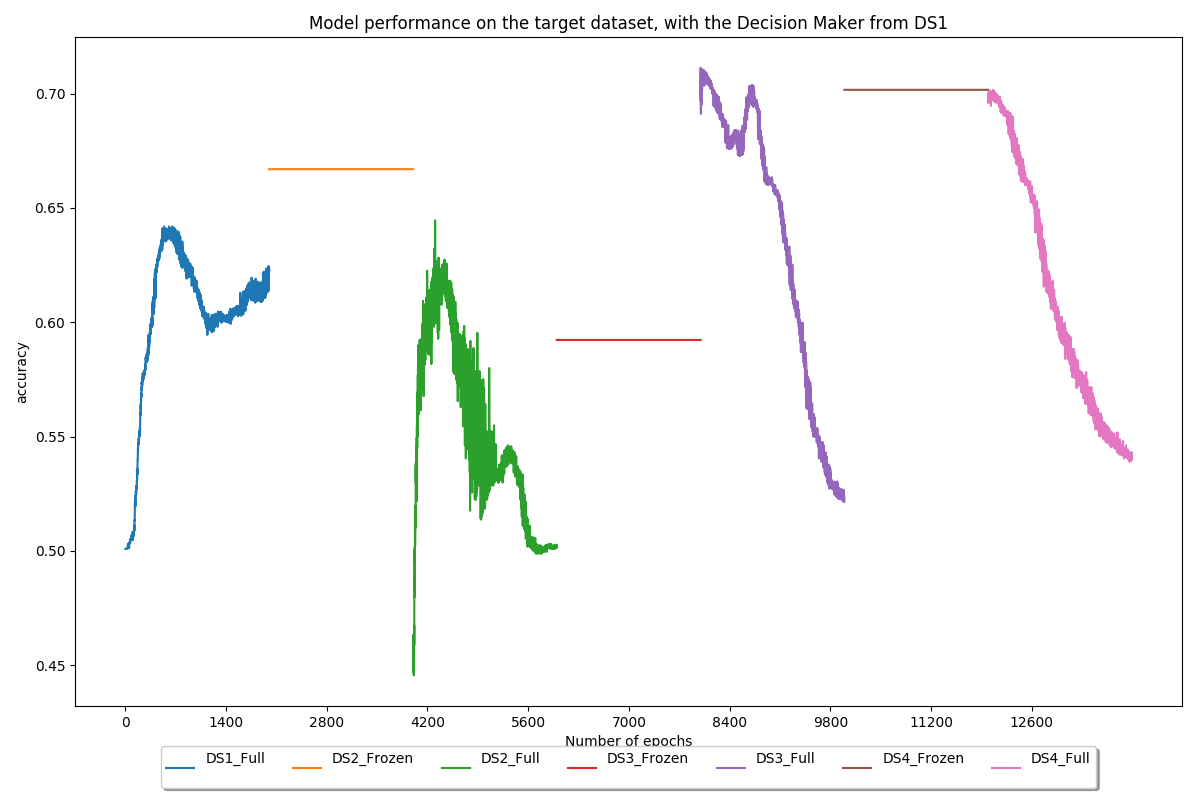
Conclusion
This work presented the process leading to the development of a deep learning system to classify potentially cancerous lesions, as well as strategies to overcome field related issues such as the lack of data. The starting point was the reproduction of Song et al.’s experiment. Reaching good performance in this part despite being able to reproduce every single trick showed that our processing and training methods were working well. This resulted in a solid baseline that was exploited in order to take part in the “SPIE-AAPM-NCI Prostate MR Classification Challenge”, also called PROSTATEx challenge. Various hyperparameters and ways of processing the data were tested in order to reach an AUC of 0.76 on this challenge. This score would have placed the model at the 15th position out of 71 submissions at the time of the challenge, which confirms the robustness of the latter.
Then, the work focused on overcoming one of the main issues in deep learning: the lack of data. To achieve this, transfer learning, as well as more common techniques such as data augmentation, were applied. Our transfer learning implementation alternated between frozen and unfrozen steps and made use of brain and lung datasets to increase the model performance on a prostate dataset. During frozen steps, the first part of the model (the feature extractor) does not update its weights at all, whereas the second part (the decision maker) does. Experiments showed that our method allowed to increase the AUC on our test set by approximately 18%, from 0.68 before transfer learning to 0.80 after transfer learning.
Throughout this work, various reusable tools were developed: visualization of medical imaging files, conversion of medical imaging files to PNG, a PyTorch sampler using undersampling, easy-to-use processing scripts for multiple datasets (PROSTATEx, Kaggle Brain, Lung CT Challenge), processing verification tools (red dot images), training verification tools (gradient flow graphs, metrics plots using Tensorboard), an all-in-one training and testing file which can be adapted to new models and datasets, an end-to-end transfer learning pipeline. All these elements can be used as a baseline for future works or as additions to existing projects. Do not hesitate to contact us for further information and source files.




