dipLODocus[RDF]
dipLODocus[RDF]
dipLODocus[RDF] is a new system for RDF data processing supporting both simple transactional queries and complex analytics efficiently. dipLODocus[RDF] is based on a novel hybrid storage model considering RDF data both from a graph perspective (by storing RDF subgraphs or RDF molecules) and from a “vertical” analytics perspective (by storing compact lists of literal values for a given attribute).
Overview
Our system is built on three main structures: RDF molecule clusters (which can be seen as hybrid structures borrowing both from property tables and RDF subgraphs), template lists (storing literals in compact lists as in a column-oriented database system) and an efficient hash-table indexing URIs and literals based on the clusters they belong to.
Figure below gives a simple example of a few molecule clusters—storing information about students—and of a template list—compactly storing lists of student IDs. Molecules can be seen as horizontal structures storing information about a given object instance in the database (like rows in relational systems). Template lists, on the other hand, store vertical lists of values corresponding to one type of object (like columns in a relational system).
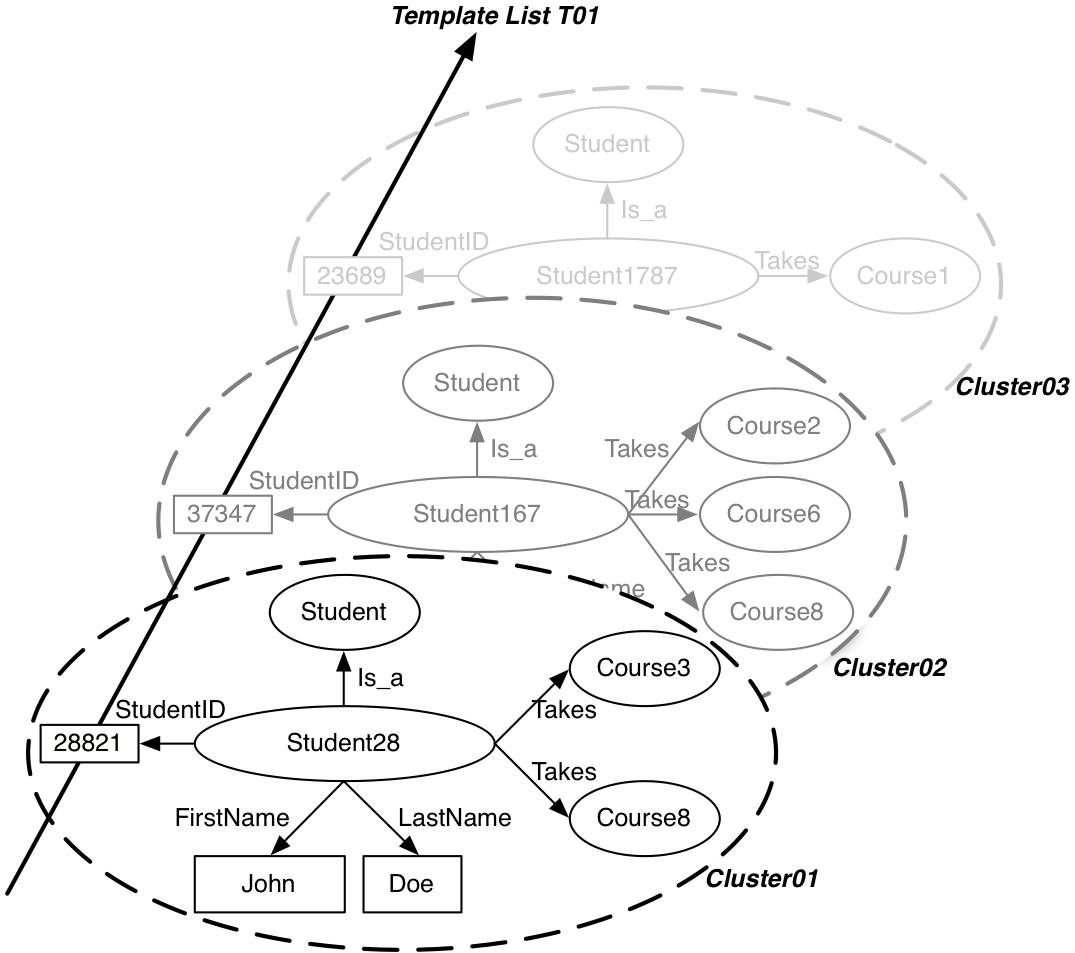
The two main data structures in dipLODocus[RDF] : molecule clusters, storing in this case RDF subgraphs about students, and a template list, storing a list of literal values corresponding to student IDs.
Architecture
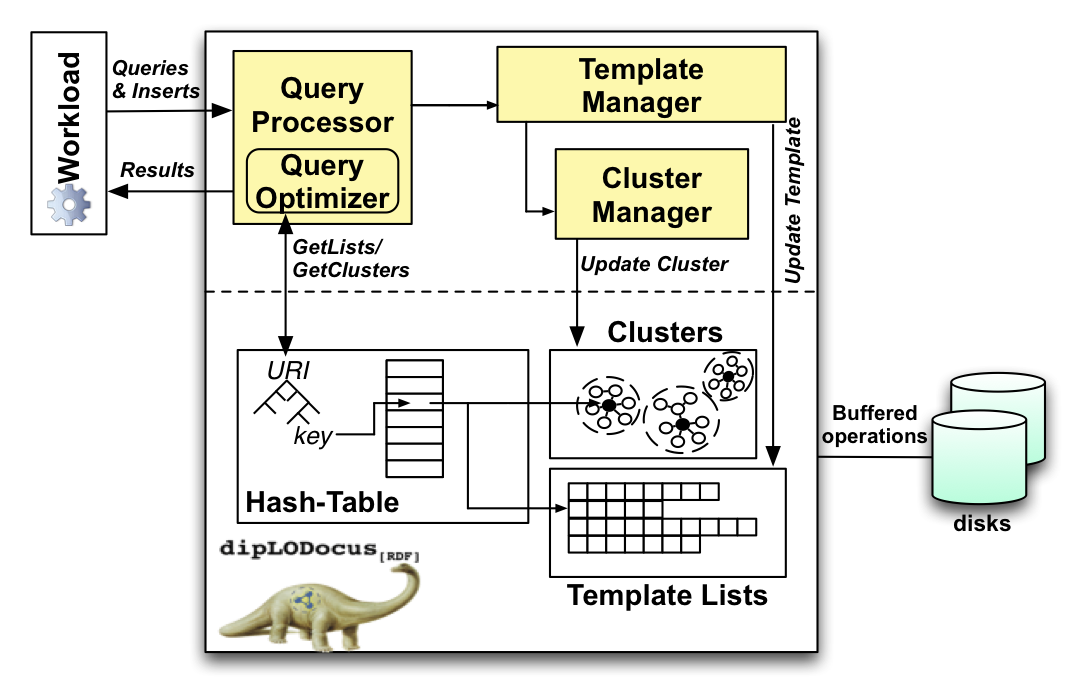
The figure below gives a simplified architecture of dipLODocus[RDF]. The Query Processor receives the query from the client, parses it, optimizes it, and creates a query plan to execute it. The hash-table uses a lexicographical tree to assign a unique numeric key to each URI, stores metadata associated to that key, and points to two further data structures: the molecule clusters, which are managed by the Cluster Manager and store RDF sub-graphs, and the template lists, managed by the Template Manager. All data structures are stored on disk and are retrieved using a page manager and buffered operations to amortize disk seeks.
The architecture of dipLODocus[RDF]:
Benchmark
We compared our prototype implementation of dipLODocus[RDF] to five other well-known database systems: Postgres, AllegroGraph, BigOWLIM, Jena, Virtuoso, and RDF 3X. We chose those systems to have dierent comparison points and because they were all freely available on the Web.
The benchmark we used is one of the oldest and most popular benchmarks for Semantic Web data called Lehigh University Benchmark. We used two data sets, the first one consisting of ten LUBM universities, and the second regrouping one hundred universities.
We compared the runtime execution for LUBM queries and for three analytic queries inspired by an RDF analytic benchmark we recently proposed (the BowlognaBench benchmark).
The three additional analytic/aggregate queries that we considered are as follows:
- a query returning the professor who supervises the most Ph.D. students
- a query returning a big molecule containing all triples within a scope of 2 of Student0 and
- a query returning all graduate students.
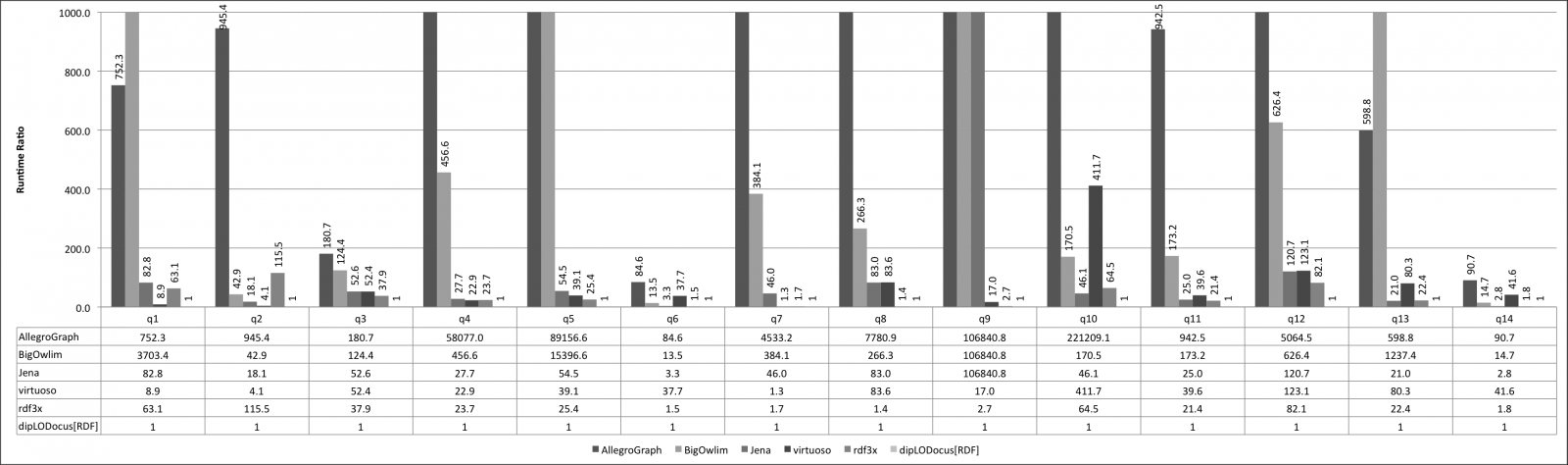
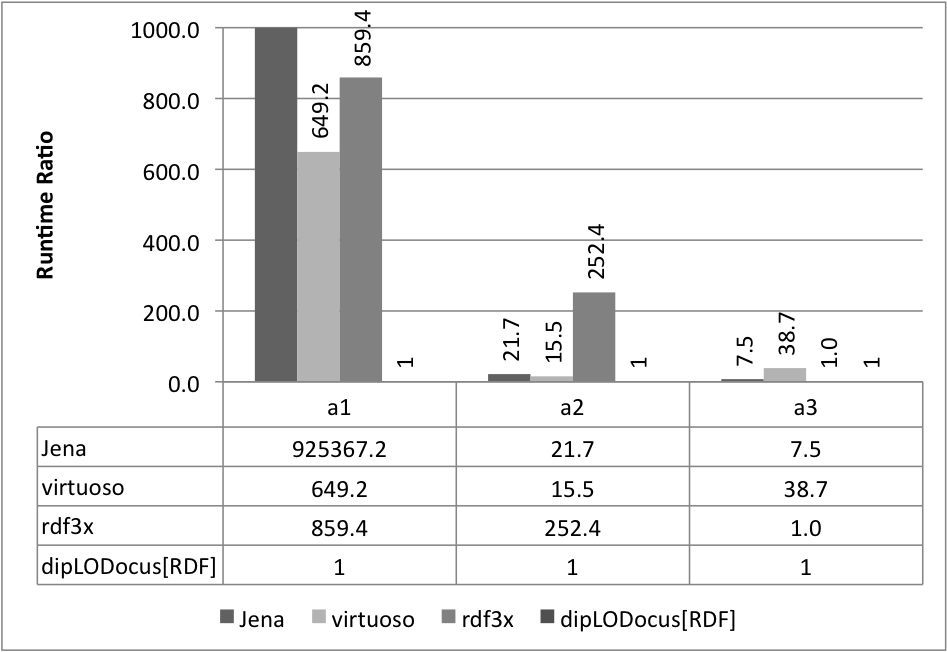
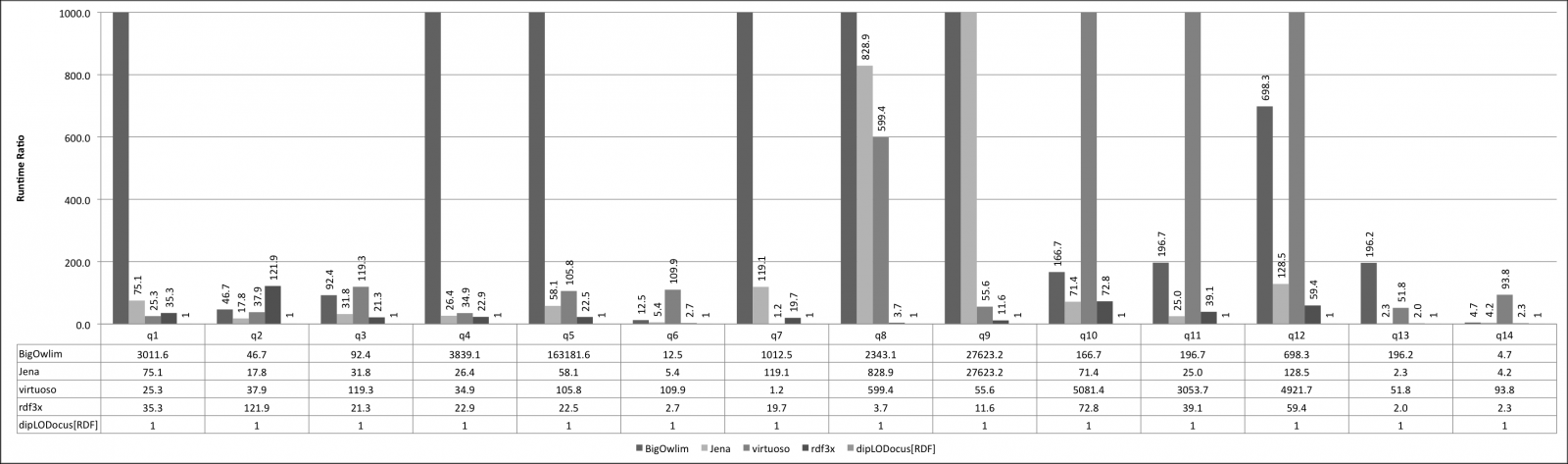
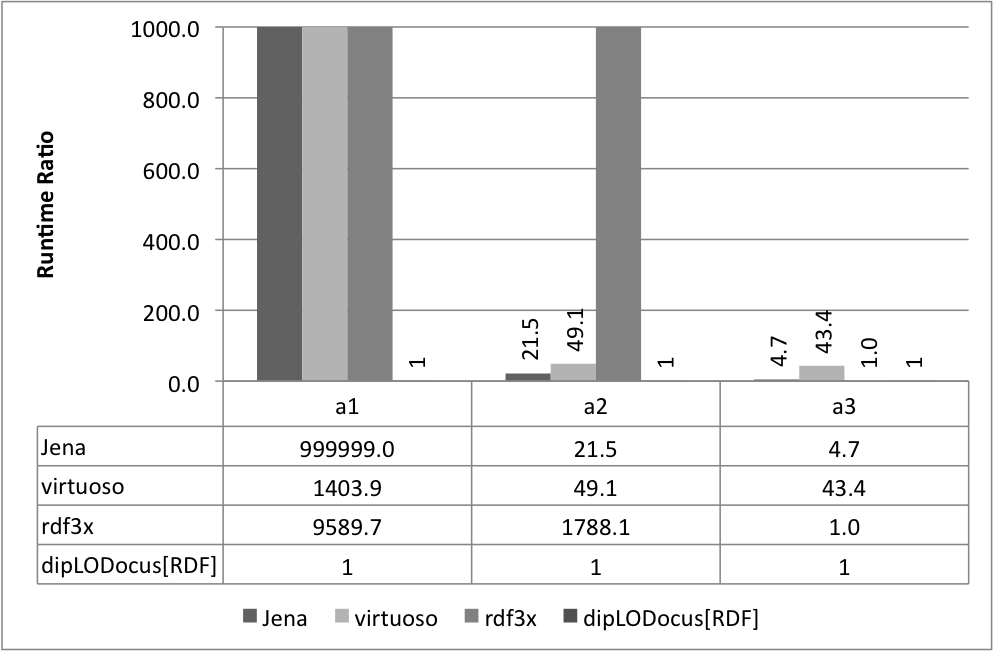
Relative execution times for all queries and all systems are given below. Results are given as runtime ratios, with dipLODocus[RDF] taken as a basis for ratio 1.0 (i.e., a bar indicating 752.3 means that the execution time of that query on that system was 752.3 times slower than the dipLODocus[RDF] execution).
LUBM queries 10 Universities:
Analytic queries 10 Universities:
LUBM queries 100 Universities:
Analytic queries 100 Universities:
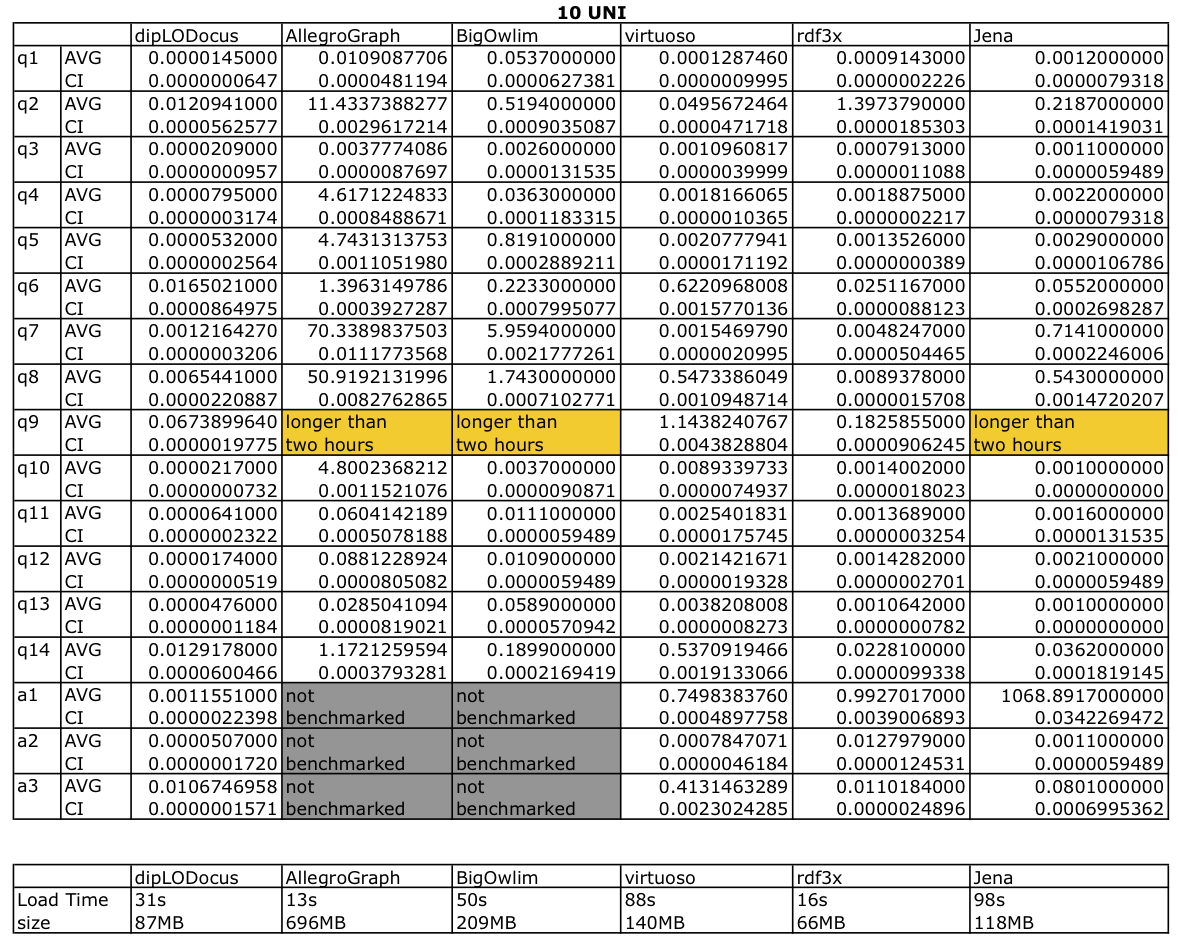
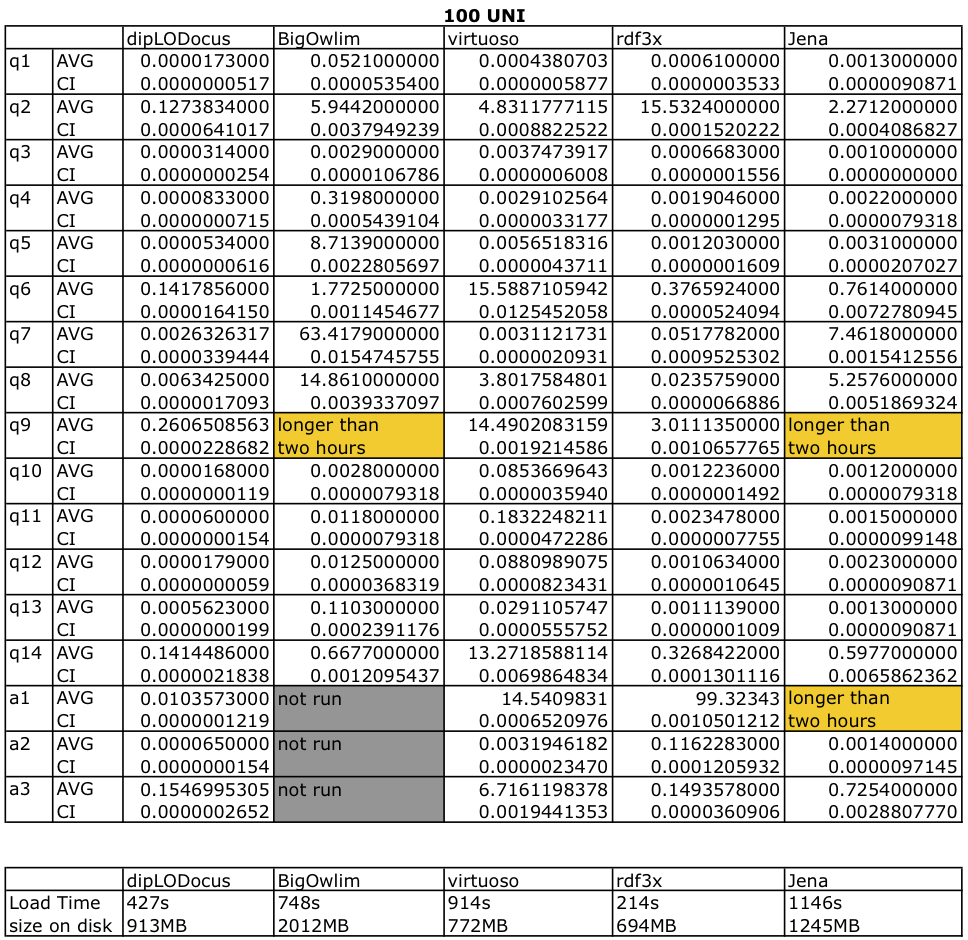
Tables bolow give execution absolute times with condence intervals at 95%, database sizes on disk and load times are given in for both datasets.
Query execution time for 10 Universities:
Query execution time for 100 Universities:
Publications
- “dipLODocus[RDF] | Short and Long-Tail RDF Analytics for Massive Webs of Data” Marcin Wylot, Jige Pont, Mariusz Wisniewski, and Philippe Cudre-Mauroux, ISWC 2011 [acceptance rate 19%], [pdf]